Products / Application / Long-Read Sequencing
Products
Nucleic Acid Extraction Kits
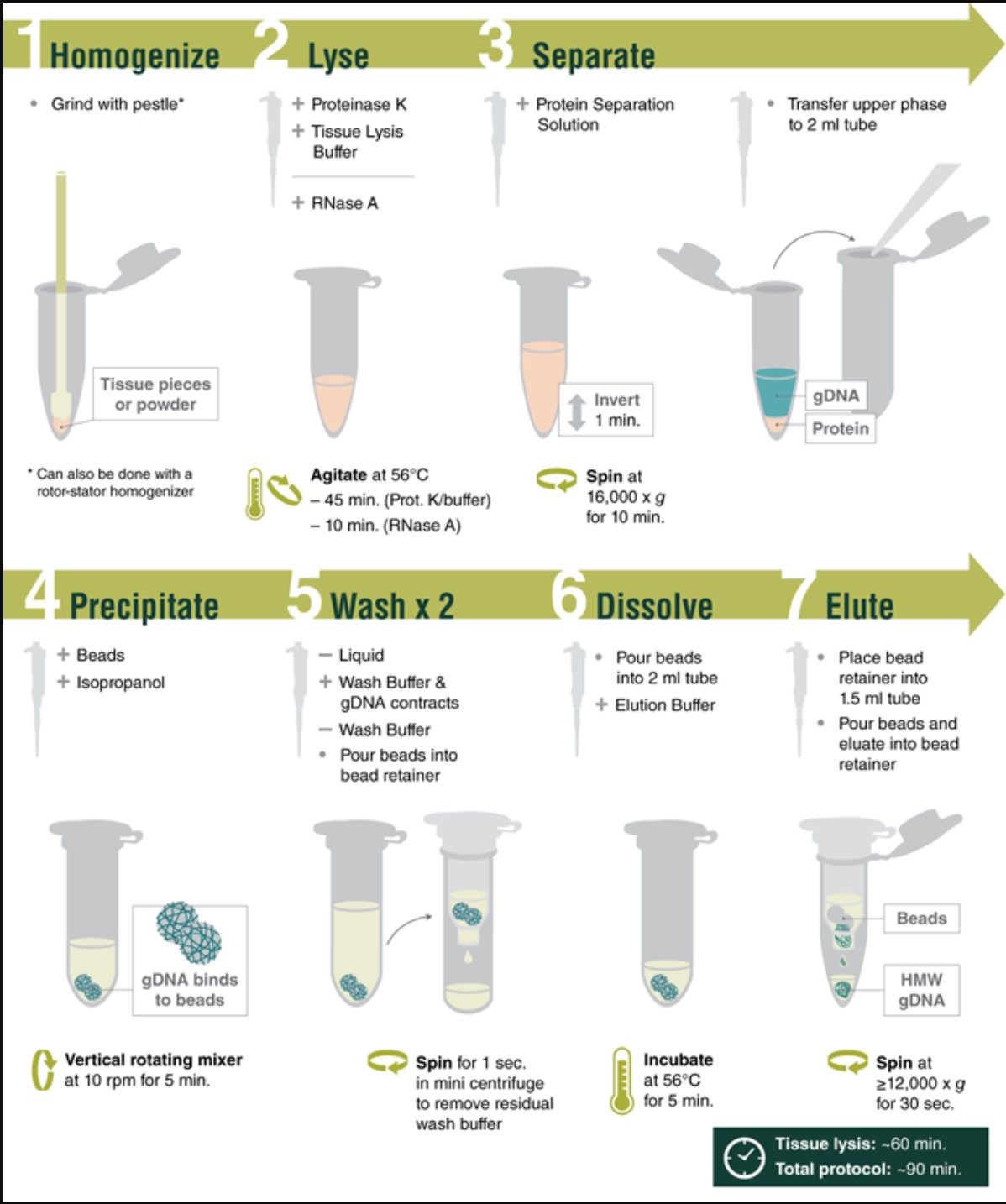
- Extremely fast, user-friendly protocols utilizing a novel glass-bead-based approach
- Reproducibly purify high molecular weight genomic DNA (HMW DNA) from various sample types
- Tune fragment length by varying agitation speeds during lysis; achieve DNA in the Megabase size range with low speeds
- Achieve outstanding results when compared to other commercially available solutions
- Excellent performance in long read sequencing
More Information:
- High Molecular Weight DNA Extraction – Product comparison
- Nucleic Acid Purification – Product overview
Microbiome DNA Enrichment
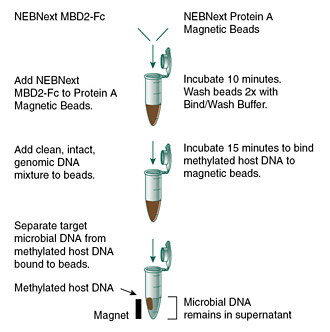
Enriching microbial DNA from samples containing methylated host DNA (including human), by selective binding and removal of the CpG-methylated host DNA.
- Fast, simple protocol
- Enables microbiome whole genome sequencing, even for samples with high levels of host DNA
- Compatible with downstream applications including next generation sequencing on all platforms, qPCR and end point PCR
- Suitable for a wide range of sample types
- No requirement for live cells
- Optional protocol to retain separated host DNA
- Also effective for separation of organelle DNA (e.g. mitochondria, chloroplast) from eukaryote nuclear DNA
More Information:
Whole Genome Amplification
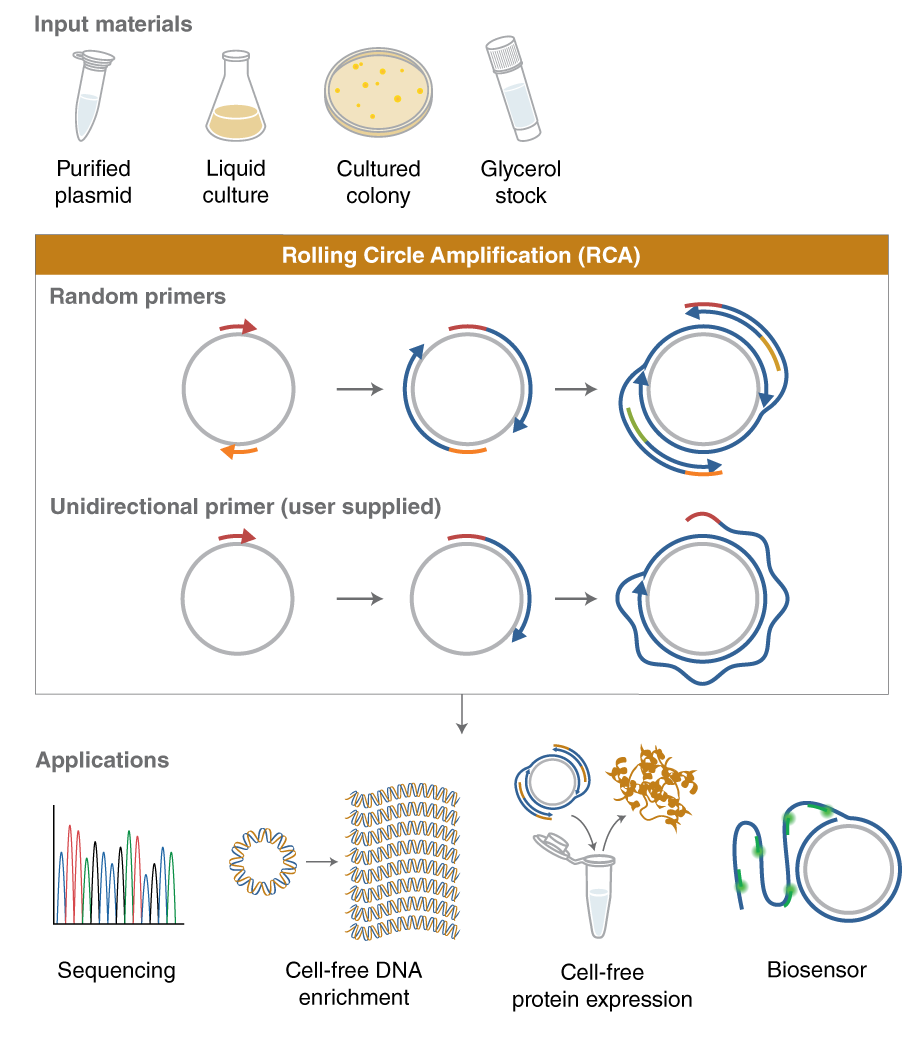
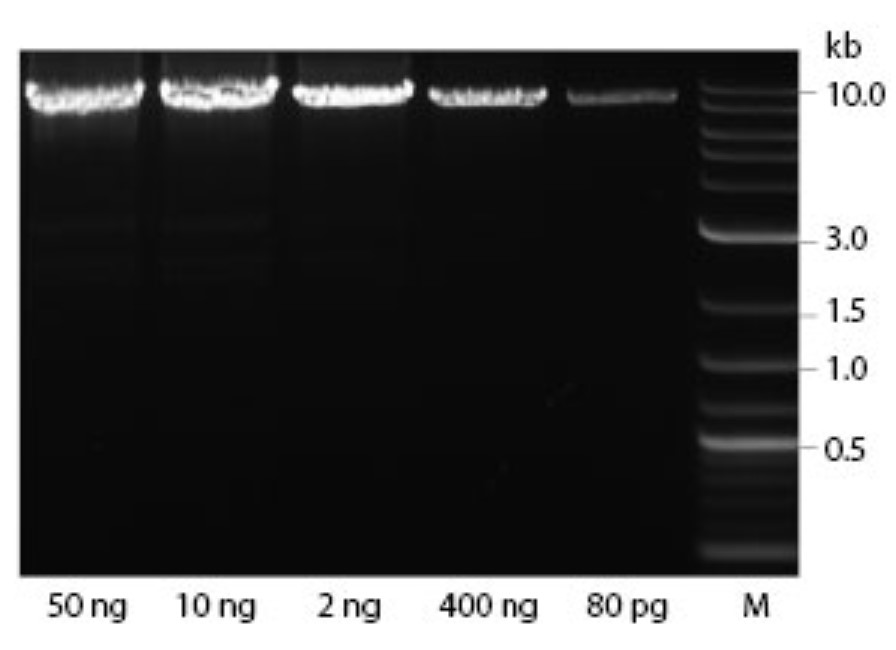
Includes everything needed for sensitive and robust rolling circle amplification (RCA), an isothermal amplification method to continuously amplify circular DNA by generating long, repetitive copies of the circular sequence.
- Powered by phi29-XT DNA Polymerase, an engineered polymerase that generates high product yield in a short reaction time with improved thermostability and sensitivity
- Amplify from as little as 1 fg of circular DNA input
- Flexible protocol offers compatibility with different types of sample input material.
- RCA products can be used directly in downstream applications such as DNA sequencing, cell-free DNA enrichment, cell-free protein expression, and DNA biosensors.
- Kit includes dNTPs and exonuclease-resistant random primers
More Information:
- Isothermal Amplification – Application overview
Amplicon-Based Enrichment
- Aptamer-based inhibitor enables room temperature reaction setup
- Optimized enzyme blend for amplification up to 30 kb
- 2X higher fidelity than Taq
- Master mix is a 2X concentrated solution containing everything needed for robust amplification of complex or simple templates
More Information:
- Tips for Amplifying Large Amplicons – Featured video
Long cDNA Synthesis
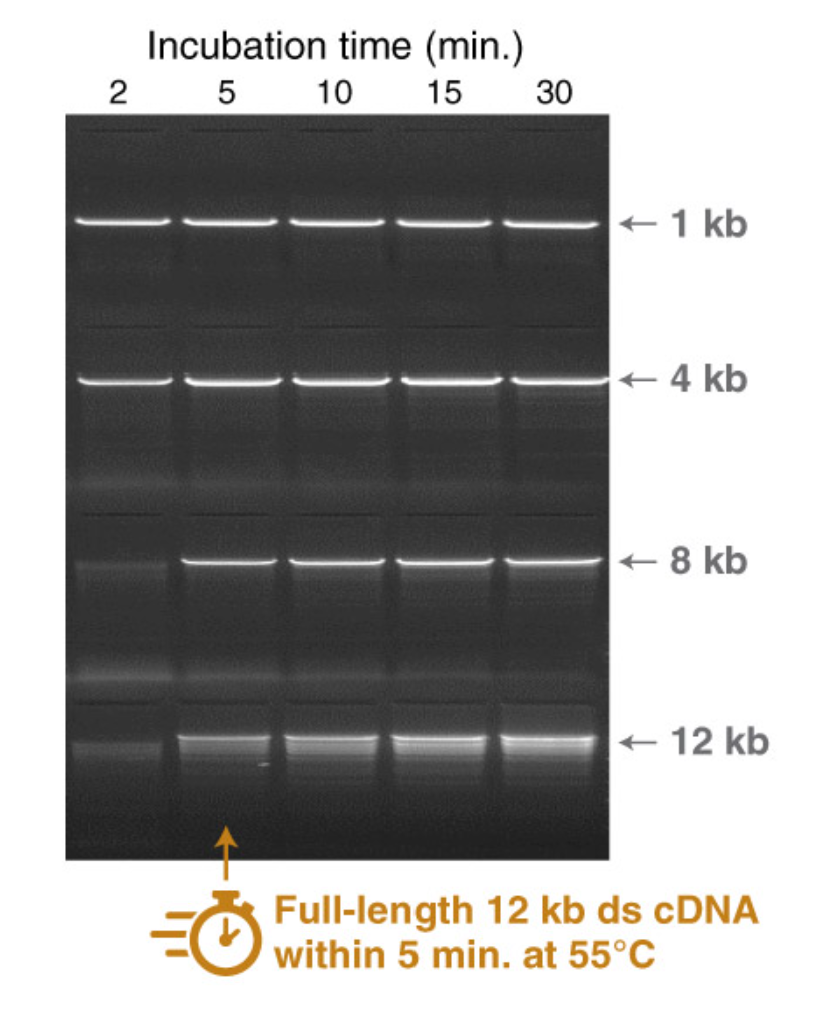
Highly processive RT enables fast reactions with high yields of long cDNA
- Strong inhibitor tolerance enables robust cDNA synthesis performance
- Supports direct RNA sequencing and long read cDNA sequencing workflows
- Increased thermostability for robust cDNA synthesis at high temperatures
- Experience comparable fidelity to retroviral RTs
More Information:
- Induro NEB TV Webinar
- Induro™, a Novel Reverse Transcriptase for Nanopore Direct RNA Sequencing with Significantly Improved RNA 5’ Coverage - Scientific poster from London Calling
Library Preparation for Oxford Nanopore Technologies

The NEBNext DNA repair, end repair and ligation reagents recommended in Ligation library preparation are now available in the same product
- Component volumes tailored for use with many SQK-LSK109, SQK-LSK110 and SQK-LSK114 workflows
- Simplified ordering and inventory management
- Compatible with all devices: MinION®, GridION®, PromethION™, Flongle®
- No more waste – no unnecessary buffers or excess reagents
More Information:
- Infectious Diseases & ARTIC Sequencing – Product overview
- Supporting Infectious Disease Research and Development – Key resources
PacBio Sequencing Solutions
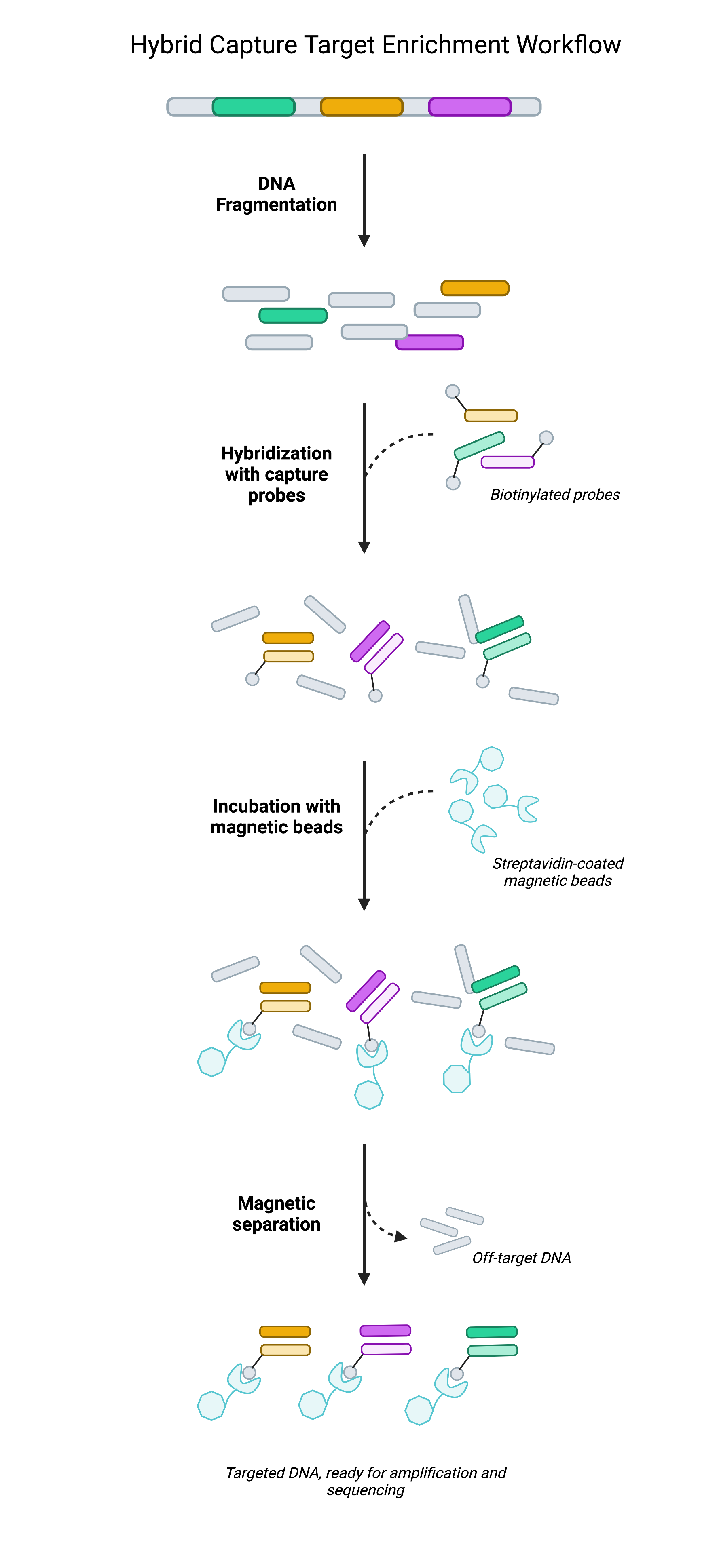

Pre-designed and custom panels are using dsDNA probes for hybridization capture-based target enrichment.
Why choosing us?
- Hybridization capture-based dsDNA probes: allows high sensitivity
- Proprietary design algorithms: design probes in order to balance the coverage across difficult-to-sequence or difficult-to-map regions
- Proprietary silicon-based DNA synthesis platform: allows industrialized the production of cost-effective, high-fidelity and high-throughput DNA
More Information:
